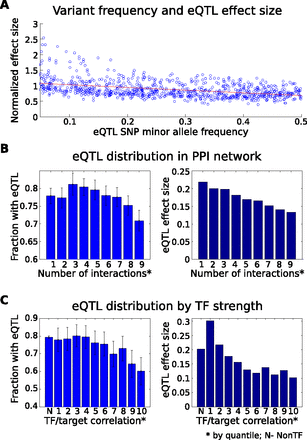
Distribution of cis-regulatory variation and selective pressure. (A) Effect size of cis-eQTLs compared to minor allele frequency of the most significant SNP per eQTL gene (computed using subsampling) (Methods). We find a strong inverse relationship (Spearman's r = −0.13, P < 10−7). If we normalize by the observed variance of each gene, the observed relationship becomes stronger (P < 10−39). (B) A depletion of cis-eQTLs is evident (P < 0.05) among genes with many protein–protein interactions (PPI); additionally, a strong negative correlation exists between the number of interactions and eQTL effect size (P < 10−35). Protein coding genes were put into quantile buckets according to the number of known PPI relationships (Methods). The fraction of genes in each bucket having a significant cis-eQTL was computed along with the average effect size for the observed eQTLs. Fewer eQTLs are observed among genes with the most interactions (hub genes). Genes in the bottom 20% may be moderately depleted as well, although confidence intervals (95%) are overlapping with the intermediate deciles. (C) The fraction of genes with a significant cis-eQTL and average eQTL effect size are shown according to an estimate of the genes' regulatory impact. Known regulatory genes were put into quantiles according to the strength of correlation observed between their expression profile and the expression of all nonregulatory genes. Nonregulatory genes are shown in the leftmost bar for comparison. Strong regulatory genes show significant depletion of eQTLs (P < 10−2) compared to nonregulators and weak regulatory genes and, similarly, reduced eQTL effect sizes (P < 10−100).











