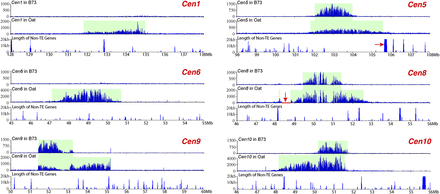
Expansion of Cen1, Cen5, Cen6, Cen8, Cen9, and Cen10 of B73 in the genetic background of oat. The top panel of each centromere shows the ChIP-seq read distribution in B73. CENH3 binding was detected in Cen5, Cen8, Cen9, and Cen10, but not in Cen1 and Cen6. The middle panel of each centromere shows ChIP-seq read distribution in the oat background. The ChIP-seq read number (y-axis) in both panels was calculated in 10-kb windows per million reads. The bottom panel of each centromere shows the length (kb) of non-TE genes in 20-kb sliding windows (step = 10 kb). The red arrow in Cen5 points to a 136.8-kb transcribed gene flanking the long-arm boundary of the CENH3-binding domain. The horizontal red bar (indicated by a red arrow) in Cen8 is a region that completely lacks ChIP-seq sequence reads. A deletion may have occurred in this region during the transfer of this maize chromosome into oat. The expansion of Cen8 surpassed this deletion.











