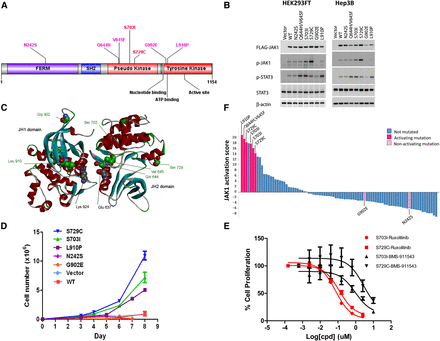
Activating mutations in Janus kinase 1 (JAK1). (A) Seven distinct somatic mutations in JAK1 in the context of protein domains and active sites. Two mutations (S703I and S729C) are recurrent, found in two samples each. (B) HEK293FT and Hep3B cells were transiently transfected with either empty vector, Flag-tagged JAK1 WT, or Flag-tagged JAK1 variants for 48 h, serum-starved for 4 h, and in the case of Hep3B, treated with vehicle or 10 ng/mL IL6 for 15 min. Resultant JAK1 and STAT3 activation determined by immunoblotting lysates with anti-pJAK1 (Y1034/1035) and -pSTAT3 (Y705), respectively. Comparable expression of Flag-tagged JAK1 constructs and protein loading was confirmed using anti-M2 Flag and -beta actin. Representative result is shown in two to six independent experiments. (C) A model of the interaction between the JAK1 JH1 and JH2 domains was generated using the crystal structure of the JH1 domain (3EYG) and a homology model of the JH2 domain built using Discovery Studio (accelrys.com). The two residues reported to be in contact (Lys 924 and Glu 637) that were used to orient the domains are highlighted (gray CPK), as are the seven residues found to be mutated in this study (green CPK), and the small-molecule CP-690,550 bound in the JH1 ATP site (ball-and-stick). (D) Tumor samples are ranked by JAK1 activation scores based on independently derived JAK1 gene expression signature (Flex et al. 2008). Colors indicate JAK1 mutation statuses and whether the mutation is activating in vitro as experimentally determined. (E) Wild-type JAK1 or activating JAK1 mutants (S703I, S729C, and L910P) transduced Ba/F3 cells were cultured in the absence of IL3 in triplicate. Cell numbers were counted at indicated time points and data presented as mean ± SD. (F) JAK1 activating mutant (S703I and S729C)-transduced cells were treated with either ruxolitinib or BMS-911453 for 3 d. Cell viability was measured and data presented as mean ± SD.











