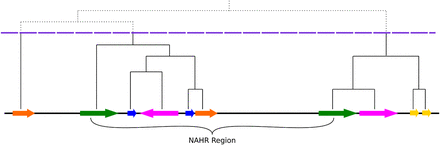
Schematic representation of LCR clustering. Horizontal arrows indicate LCR elements and their orientation; the same color represents a pair of paralogous LCRs. A hierarchical clustering tree is depicted above; the dashed horizontal line (violet) shows the height threshold for cutting this tree. Directly oriented paralogous LCRs (DP-LCRs) can potentially mediate NAHR events. The structure of LCR clusters (subunit structure, orientation, etc.) as well as the DNA sequence homology between LCR clusters flanking NAHR-prone regions often revealed extensive complexity, in contradistinction to the concept of a “segmental duplication” and more consistent with “complex LCR clusters” and with current accepted models for generating duplications and complex genomic rearrangements; e.g., FoSTeS (Lee et al. 2007) or MMBIR (Hastings et al. 2009) (Supplemental Fig. S1).











