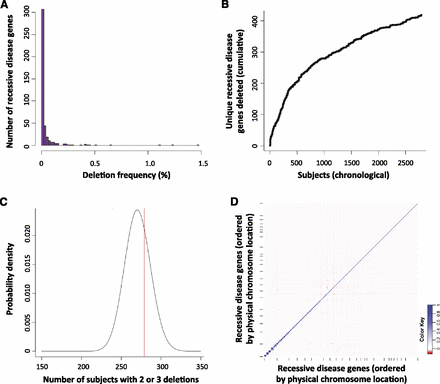
Allele frequency spectrum and assortment of heterozygous Tier 1 deletions (potential carrier CNVs). (A) Histogram of the prevalence with which each of 419 recessive disease genes is deleted by heterozygous Tier 1 CNVs, demonstrating a predominance of rarely affected genes and a few more commonly deleted genes. (B) Chronological ascertainment of unique recessive disease genes affected by heterozygous Tier 1 deletions. As more individuals with Tier 1 heterozygous deletions are analyzed (x-axis), additional recessive disease genes are identified that were not previously found to be deleted in our cohort, up to a total of 419 of 1228 known recessive disease genes (34%). The ascertainment of unique, deleted recessive disease genes continues to rise even after assessing 2829 subjects with a Tier 1 heterozygous deletion. (C) To determine whether Tier 1 heterozygous CNVs are distributed randomly among subjects, we compared the number of V8 individuals with two or three Tier 1 heterozygous CNVs deleting a single recessive disease gene (279 subjects; red line) to that expected by chance (black probability distribution; see text and Supplemental Methods). There was no statistically significant enrichment of individuals with multiple potential carrier deletions (P = 0.312), suggesting that carrier CNVs, numerically, are distributed randomly among our cohort. (D) Co-occurrence (or absence of co-occurrence) of heterozygous deletions in all pairs of 374 recessive disease genes among V8 cases displayed as a correlation matrix. Genes are plotted along each axis consecutively by genomic position. (Blue) Relative enrichment of codeletion; (red) relative paucity of codeletion.











