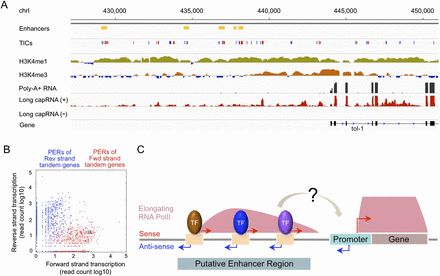
Definition of a novel regulatory architecture. (A) Genome browser image of signals indicating enhancers, TICs (red: forward strand; blue: reverse strand), H3K4me3, H3K4me1, whole-cell (mainly cytoplasmic) RNA, and nuclear long cap RNA from the plus and minus strands, with gene annotation below. (B) RNA Pol II elongation across enhancers is in the same orientation as that of the nearest downstream gene. Plots show forward strand long cap RNA-seq read count (log10 scale) relative to reverse strand signal over enhancers upstream of forward (red) or reverse (blue) strand tandem genes. The red dots showing enhancers upstream of the tol-1 gene (forward strand) shown in A were circled. (C) Proposed model illustrating the architecture and potential regulatory roles of transcribed enhancers.











