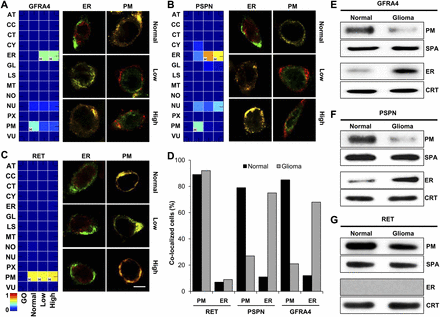
Conditional location of GFRA4, PSPN, and RET in glioma. (A–C) CLMs of GFRA4 (A), PSPN (B), and RET (C), and the results of confocal images in normal brain and glioma tissues. The color of the heat maps indicates predicted degree of possibility, and “H” indicates the location with the highest degree of possibility within each condition. Scale bar, 5 μm. (D) Location fraction of GFRA4, PSPN, and RET in normal brain and glioma primary cells. (E–G) Results of cellular subfractionation and Western blotting for locations of GFRA4 (E), PSPN (F), and RET (G) in normal brain and glioma primary cells. (SPA) sodium potassium ATPase (plasma membrane marker), (CRT) calreticulin (ER marker).











