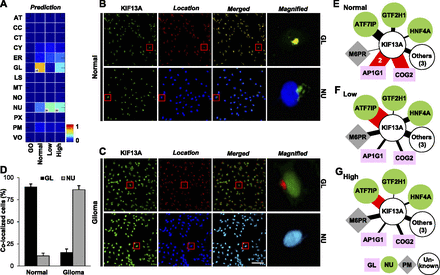
Prediction and validation of KIF13A mislocation in glioma tissues and cells. (A) Conditional location map for KIF13A, for which the highest signal under normal conditions was in the Golgi apparatus (GL), but in low- and high-grade gliomas was in the nucleus (NU). The color indicates degree of possibility and “H” indicates the location with the highest degree of possibility within each condition. (B,C) Confocal images for KIF13A (green) together with markers for GL (red, row 1) or NU (blue, row 2) in normal (B) and glioma (C) tissues reveals results consistent with predictions. Scale bar, 5 μm. (D) The fraction of colocalized cells expressing GL and NU markers using >50,000 normal brain and glioma primary cells. Samples from four normal and five glioma subjects were used. (E–G) The dynamic interaction neighborhood of KIF13A in normal brain (E), and low- (F) and high-grade (G) glioma tissues. Node color/shape indicates known protein locations. The width of each link is proportional to the expression coherence score, which is also indicated numerically for selected links. Red links indicate key interactions.











