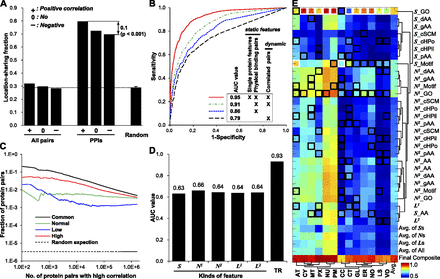
Models generated and usefulness of coherent protein interactions for location prediction. (A) The percentage of protein pairs sharing at least one location, calculated from different sets of proteins. “Random” was calculated as the average of 1000 randomly selected interaction sets with the same number of interactions as the original protein network. (B) Leave-two-out cross-validation with a DC-kNN classifier was used to assess the effect of static and network features on the accuracy of predicting known subcellular locations. (C) Fractions of protein pairs with known interactions among the top-k pairs with highest correlations in expression. “Common” indicates pairs common to normal brain (Normal) and low (Low)- and high (High)-grade gliomas. (D) Average AUC values of different feature sets, including S, ND, and LD. Here, the “TR” category means the final average AUC value of the selected models for 13 locations in the training stage. “D” indicates the distance of incorporated network neighbors. (E) Generated models with selected feature sets for individual locations using a DC-kNN classifier. Black and white squares represent selected feature sets for each location, with the white square denoting the best feature set overall. The last row indicates the AUC values for prediction of individual locations. The last column indicates the average AUC values of individual feature sets across the 13 locations considered.











