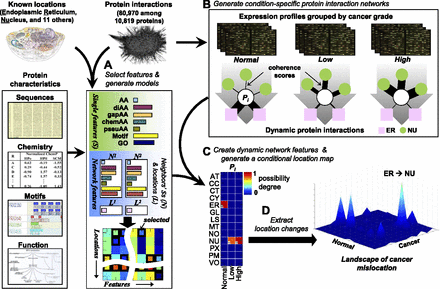
Proteome-wide prediction of protein mislocation. (A) A protein is described by its sequence, chemical properties, motifs, and functions (single protein features) together with a description of its network neighborhood (capturing the features of its neighbors and their subcellular locations, if known). The best combination of features for each location is selected using a DC-kNN classifier. (B) Condition-dependent dynamic network features are generated by assigning different weights to each neighbor of a protein, depending on their similarity in gene expression profiles. (C) Selected features from A are combined with condition-dependent networks from B to compute a CLM for the protein, listing the quantitative possibility that the protein is in each location under each condition. (D) Mislocations are identified by calculating differences in degrees of possibility across conditions.











