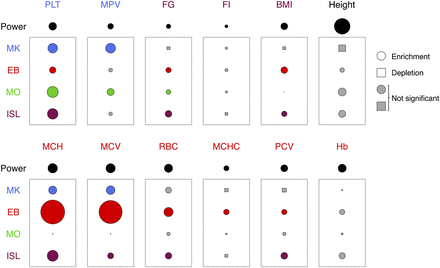
Enrichment patterns of quantitative trait-associated variants with small effect sizes at cell type–restricted NDRs. The data points shown as circles and rectangles represent the deviation of the P-value distribution of SNPs at NDRs restricted to MKs, EBs, MOs, or pancreatic islets (ISLs) from the P-value distribution of matched randomly sampled SNPs at the 0.005 quantile (Supplemental Fig. 8). Thus, this deviation measures the level of enrichment of associated sequence variants at NDRs, where the circle and rectangle surface areas represent level of enrichment (mean ratios > 1) and depletion (mean ratios < 1), respectively. Gray symbols represent ratios that are not significantly different from 1; i.e., the mean ratio across replicates was within 2 SDs of 1. The level of enrichment is indicated for sequence variants associated with two platelet traits ([PLT] platelet count; [MPV] mean platelet volume), six erythrocyte indices ([Hb] total hemoglobin concentration; [PCV] packed red cell volume; [RBC] red blood cell count; [MCHC] mean red cell hemoglobin concentration; [MCH] mean red cell hemoglobin; [MCV] mean red cell volume), as well as four nonhematological quantitative traits ([FG] fasting glucose; [FI] fasting insulin; [BMI] body mass index; height). The circle area labeled “Power” gives a quantification of the amount of signal present in each GWA data set. Specifically, it represents the deviation of the P-value distribution of all tested SNPs from the expectation under the null at the 0.005 quantile.











