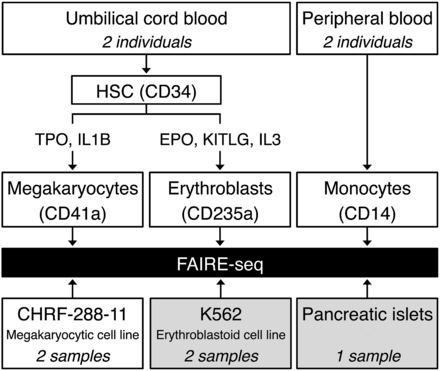
Overview of the study design. Cord blood–derived CD34+ hematopoietic progenitor cells from two unrelated individuals were differentiated in vitro into either megakaryocytes (MKs) or erythroblasts (EBs). Monocytes (MOs) were purified from peripheral blood from another two individuals. We also prepared FAIRE samples from CHRF-288-11 megakaryocytic cells. In addition, we retrieved publicly available FAIRE-seq data sets for K562 erythroblastoid cells and pancreatic islets from The ENCODE Project Consortium (2012) and Gaulton et al. (2010), respectively, and reanalyzed the data sets in concordance with all other FAIRE data sets. (HSC) Hematopoietic stem cell; (TPO) thrombopoietin; (IL1B) interleukin 1, beta; (EPO) erythropoietin; (KITLG) KIT ligand (also known as SCF, or stem cell factor); (IL3) interleukin-3.











