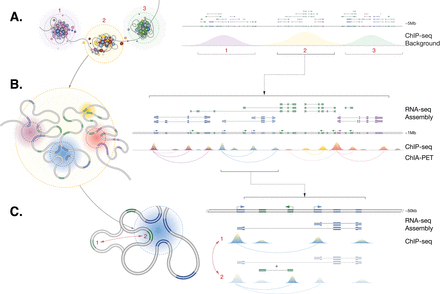
Three-dimensional interpretation (left) of regulatory and transcriptional complexity in one-dimensional genome representation (right). (A) The genome forms large complex clusters and introspective folded clusters with specialized transcription compartments. Each of these clusters correlates to a collection of transcripts and “background” ChIP-seq enrichment. (B) Within each cluster the genome is folded to associate with subnuclear structures containing transcription factors and machinery, splicing, and other accessory proteins. These associations coregulate genes to generate interleaved complex transcriptional networks of coding (blue) and noncoding transcripts (green). Proximal cross-linking with ChIP-seq results in a complex landscape of enrichment across loci that reflect the folded genome structure. (C) Within each gene, local dynamic chromatin folding determines the association of alternative promoters and local noncoding RNAs with a shared regulatory architecture, thereby mediating coregulated gene expression.











