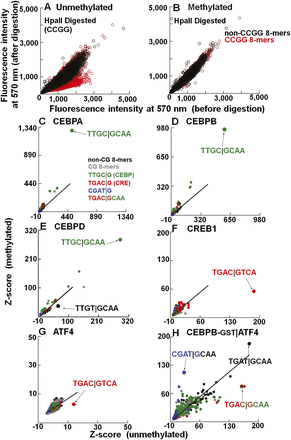
The effects of methylation on DNA binding properties of B-ZIP proteins. (A,B) Validation of CG methylation using methylation-sensitive (HpaII) endonuclease. The 40K feature microarray is scanned at 570 nm to detect Cy3-cytosine spiked into the DNA double-stranding reactions. Fluorescence intensities before and after methylation were normalized. DNA features containing CCGG are in red. Fluorescence intensities on (A) unmethylated and (B) methylated arrays before and after HpaII digestion. (C–H) Z-scores for all 32,896 8-mers from unmethylated and methylated 40K feature microarray. Lines are fitted to the non-CG 8-mers, which show no change in Z-scores between unmethylated and methylated arrays and serve as an internal control. The 8-mers are color-coded: CG (gray), non-CG (black), TTGC|G (green), TGAC|G (red), CGAT|G (blue), and chimeric CRE|CEBP sequence TGAC|GCAA is shown in brown. The best-bound 8-mers are indicated by arrows. (C) CEBPA-GST. (D) CEBPB-GST. (E) CEBPD-GST. (F) CREB1-GST. (G) ATF4-GST. (H) CEBPB-GST|ATF4.











