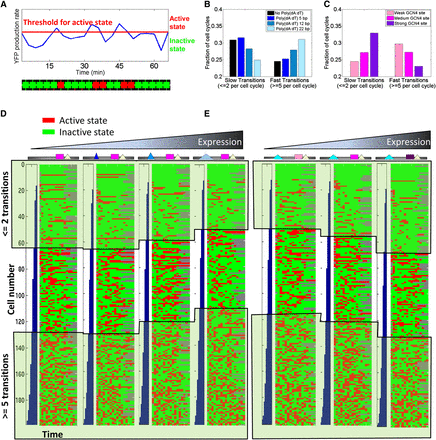
Lengthening poly(dA:dT) tracts and strengthening transcription factor binding sites have opposing effects on the rate of promoter transitions between active and inactive states. (A) Illustration of our analysis of promoter transition rates. For each cell cycle of every cell, we classify its trace of normalized YFP production rate (blue trace illustrated here for one cell cycle of one cell) into active (red) and inactive (green) states according to whether they are above or below a predefined arbitrary threshold (red horizontal line), respectively. (B) Increasing the length of a poly(dA:dT) tract results in a higher rate of transitions between active and inactive states. For promoter variants that differ in the length of a poly(dA:dT) tract, shown is the fraction of all of its measured cell cycle traces in which the number of transitions between active and inactive states was at most two (slow transitions; left bar graph) or at least five (fast transitions; right bar graph). The comparison of these different promoter variants was done at a threshold in which the fraction of all inactive states in each variant was 70% (since absolute expression levels vary across variants, the absolute threshold value is different for each variant). See Supplemental Figure S4 for similar analyses at a range of thresholds from 50% to 90%. (C) Increasing the affinity of a transcription factor binding site results in a lower rate of transitions between active and inactive states. Same as B, but for variants that differ in the affinity of a Gcn4 binding site. (D) Visual illustration of cell cycle traces corresponding to the bar graphs from B in which the length of poly(dA:dT) tracts was varied. For each promoter variant, shown are 200 rows that each correspond to a time trace of one cell cycle of one cell with colored entries representing active (red) or inactive (green) states at a threshold in which 70% of all states were inactive. Rows are sorted according to the number of transitions between active and inactive states, and the 200 rows were sampled from all cell cycle traces such that they accurately represent the same probability distribution of the number of transitions across all cell cycle traces. (E) Same as D, but for the bar graphs from C in which the affinity of Gcn4 sites was altered.











