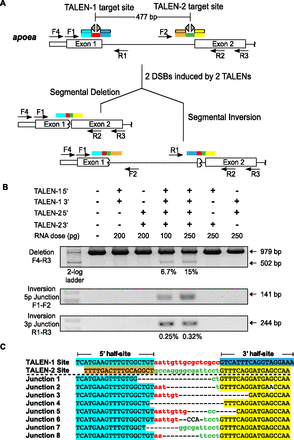
TALEN-generated deletions within the apoea locus. (A) Schematic diagram of the zebrafish apoea locus displaying the positions of the two proximal TALEN binding sites separated by ∼500 bp. The half-sites recognized by each TALEN are differentially colored (cyan, blue, orange, yellow), as are the spacers between the half-sites (red, green). Examples of the possible alterations (segmental deletions or inversions) in a single chromosome in the context of two DSBs are indicated below with the expected rearrangement of the TALEN target sites. Each primer position and directionality is indicated by an arrow within the schematic. (B) Segmental deletions are readily detected in normal-appearing zebrafish embryos when two TALENs are coinjected, where the primer pair used for detection is listed to the left of the gel image. The presence or absence of each TALEN component (5′ and 3′ recognition unit) is indicated above each gel lane, as is the dose of each component. PCR products corresponding to the unmodified locus (979 bp) and the segmental deletion (∼502 bp) are present, where the latter occurs only in embryos injected with both TALENs. Estimated deletion rates are indicated below the positive lanes. Segmental inversions are also detected in embryos treated with two TALENs, where the inversion rates are lower than the corresponding segmental deletions. (C) Junction sequences for the 500-bp segmental deletions at the apoea locus. The TALEN-1 and TALEN-2 recognition sequences are indicated above the junction sequences, where the half-sites recognized by each TALEN are differentially colored. PCR products spanning this region were shotgun-cloned and sequenced to define the junctions at these deletions. This reveals a heterogeneous set of fusion sequences between the TALEN binding sites, where microhomologies are present at many of these fusion points.











