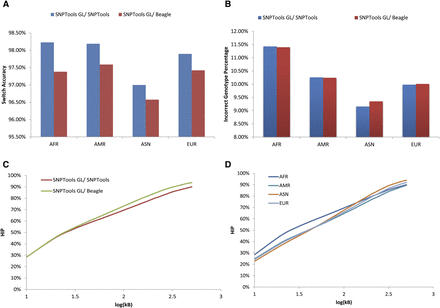
Haplotype phasing accuracy evaluation. SNPTools and Beagle are compared against the benchmark haplotypes from the 1000G Phase 1. (A) Switch accuracy between SNPTools and Beagle showed that SNPTools had higher switch accuracy. (B) While SNPTools had moderately worse performance on incorrect genotype percentage (IGP) for admixture populations (American [AMR] and African [AFR]), it showed comparable performance on all other populations. (C) Incorrect haplotype percentage (HIP) for AFR samples (representative of all populations). Phasing by SNPTools and Beagle were comparable until 100 kb [log10 (100kb) = 2]. At longer distances, SNPTools was moderately more accurate than Beagle. (D) Phasing by SNPTools on AFR, Asian (ASN), AMR, and European (EUR) populations shows that AFR samples were more likely to be incorrectly phased at a given distance (data not shown). However, at 100 kb, all populations have an HIP of 65%–70%.











