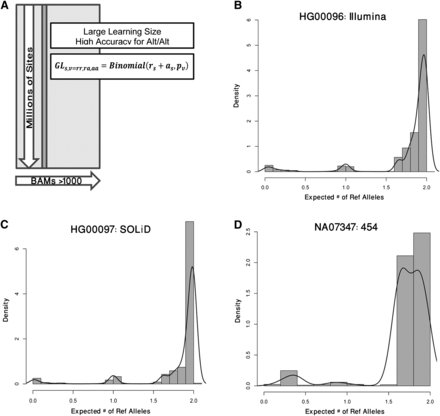
Illustration of the BBMM. (A) BBMM models each BAM as a mixture of three binomials that represent the three genotypes classes (rr = Ref/Ref, ra = Ref/Alt, and aa = Alt/Alt). Each of these classes has a class-specific binomial probability, pv which is defined as the probability of a reference read for a given genotype. BBMM estimates the parameters for each BAM by pooling data from all variant sites (approximately 34 million candidate sites that we discovered in the 1000G). (B–D) To qualitatively view the cluster assignment for each site, we compute an expected number of reference alleles by multiplying the genotype likelihood (GL) for each genotype by the number of reference alleles. We find that BBMM is able to cluster the genotypes for Illumina, SOLiD, and 454 sequencers. As representative samples, we plot HG00096 sequenced with the Illumina platform and aligned using BWA (B), HG00076 sequenced with the SOLiD platform and aligned using BFAST (C), and NA07347 sequenced by the 454 platform and aligned using SSAHA (D).











