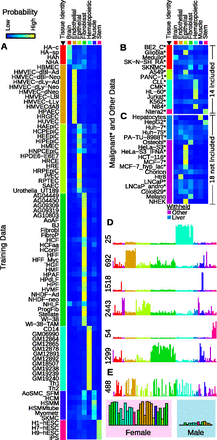
Tissue and sex classifiers based on DNase I data. Predictions from a multinomial logistic regression classifier trained to predict tissue identity for a given sample with data from 43 DHSs. (A) Predictions for training data, along with known tissue of origin (left column). Colors within the heatmaps reflect the predicted probability of belonging to each of the seven tissue classes. (B) Predictions for malignant samples not included in the training, but whose presumed tissue of origin was included in the model. (*) Malignant samples. (C) Predictions for samples whose tissue (or presumed tissue) was excluded from the training because tissue types had fewer than five samples. (D) The DNase I signal profiles of seven (out of 43) clusters selected by the model with positive coefficients. (E) The DNase I profile for the single sex-specific site (chrX:130926460–130926610) selected by the classifier. The enlarged barplot shows the distinction between samples divided by sex for the subset of samples included in the model.











