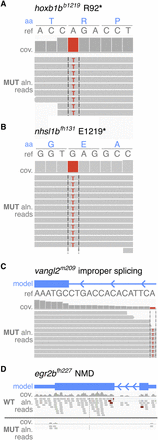
RNA-seq–based mapping identifies candidate mutations creating nonsense and missense changes, affecting splicing, and affecting gene expression. Reads are shown aligned to each known lesion site. Aligned reads are shown as gray boxes; differences from reference (ref) are highlighted by colored letters. (aa) Amino acid; (cov) coverage; (aln) aligned. (A–C) RNA-seq data from the mutant pool identified the known A-to-T transversion in hoxb1bb1219 (A), the G-to-T transversion in nhsl1bfh131 (B), both creating stop codons, and the creation of a splice acceptor introducing 15 bp of intronic sequence in the vangl2m209 mutation (C). (D) The down-regulation of egr2b (via NMD of the egr2bfh227 nonsense mutation) is evident in a comparison of the wild-type and mutant aligned reads (identified as significantly down-regulated by 25-fold via Cufflinks, q = 0.00011423).











