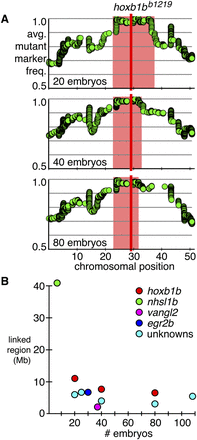
Increasing the number of embryos in an RNA-seq–based mapping experiment decreases the linkage size of the mapped region. (A) Detail of chromosome 12 containing the linked interval for each hoxb1b1219 mapping experiment. Each row is labeled with the number of embryos used in the experiment. The average frequency of mutant markers (green discs) is plotted against chromosomal position. A red box marks each region of linkage, and a red line marks the position of the hoxb1b gene; linkage was defined as the region between the “leftmost” and “rightmost” positions within 1% of homozygosity. Each y-axis is the same as in the first row. (B) Comparison of linked regions to the number of embryos used in each RNA-seq–based mapping experiment. The hoxb1b1219 experiments are labeled in red; nhsl1bfh131, in green; vanglm209, in magenta; and egr2bfh227, in blue; and unknown mutations mapped using this method, in cyan. Increasing the number of embryos decreases the linked region with diminishing returns.











