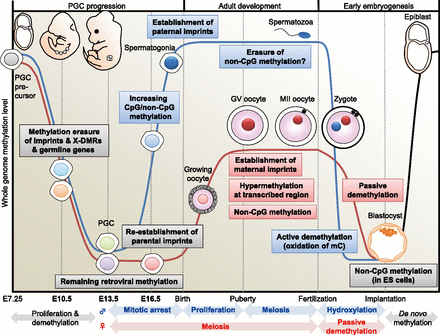
DNA methylome changes during gametogenesis and embryogenesis. Mouse PGCs emerge from precursor cells in the proximal epiblast at E7.25. They proliferate and migrate toward the genital ridge. Then, DNA methylation is globally decreased in both males (blue line) and females (red line) with erasure of methylation marks of imprinted genes, X-linked genes (only in females), and some germline-specific genes (see Fig. 4) through TET-catalyzed oxidation (Hackett et al. 2013). During this migration, whole-genome CpG methylation levels are relatively higher in males than in females (see Fig. 2). Following gonadal sex determination, new DNA methylation patterns are established in each germ cell in a sex-specific manner. In the male embryo, de novo CpG and non-CpG methylation occurs in mitotically arrested gonocytes (see Fig. 5). Establishment of the paternal methylation imprints (e.g., H19) is completed before birth (meiosis), and these imprints are maintained during subsequent spermatogenesis and throughout meiosis; however, the presence of non-CpG methylation is rarely observed in the mature spermatozoon. In the female embryo, PGCs enter meiosis as primary oocytes and arrest in the prophase of the first meiotic division; the oocyte genome remains globally hypomethylated, but parts of maternal ICRs (e.g., Peg10, Mest, Peg3, Snrpn) exhibit partial methylation (see Fig. 4). DNA methylation marks are established after birth during the growth phase of the oocyte. At puberty, fully grown oocytes are still arrested at meiotic prophase, a stage known as the germinal vesicle (GV) stage. The GV oocyte genome exhibits global hypermethylation at transcribed regions, but the whole-genome CpG methylation level of oocytes is less than half that of spermatozoa (Kobayashi et al. 2012). When GV oocytes resume the first meiotic division, they undergo GV breakdown, extrude a first polar body, and develop to metaphase of the second meiotic division (MII). MII oocytes complete meiosis only with fertilization. In the zygote, striking asymmetric DNA demethylation between the two parental genomes is observed within the zygote's cytoplasm. The paternal genome is actively demethylated before the first mitotic division through the involvement of TET protein-mediated 5-methylcytosine oxidation (conversion to 5-hydroxymethylcytosine) (Gu et al. 2011; Wossidlo et al. 2011). The maternal genome resists hydroxylation (Nakamura et al. 2012) and instead undergoes passive DNA replication-dependent demethylation. Multiple maternal and zygotic DNA-binding factors specifically recognize ICRs and protect them from these post-fertilization demethylation events (Nakamura et al. 2007; Li et al. 2008; Messerschmidt et al. 2012), resulting in epigenetic allelic asymmetries that affect associated imprinted genes. Following blastocyst implantation, the embryo undergoes a wave of de novo methylation (black line) that establishes a new DNA methylation landscape, and this process is associated with cellular differentiation.











