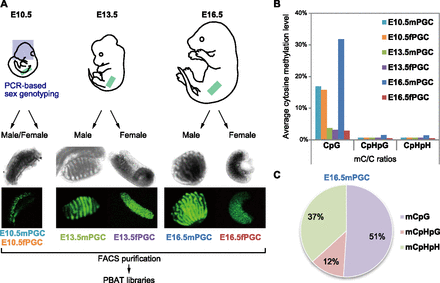
Scheme of mouse developmental-germline DNA methylome analyses. (A) Morphology of Pou5f1-ΔPE-GFP PGCs in male and female fetal gonads at E10.5, E13.5, and E16.5. The sexes of embryos at E10.5 were determined by the presence or absence of the Zfy gene. At E13.5 and E16.5, male gonads could be distinguished by the presence of testicular cords. (B) Whole-genome methylation levels (mC/C ratios) in each PGC at CpG, CpHpG, and CpHpH sites (H = C, T, or A). (Aqua bars) E10.5mPGC; (orange bars) E10.5fPGC; (green bars) E13.5mPGC; (purple bars) E13.5fPGC; (blue bars) E16.5mPGC; (red bars) E16.5fPGC. (C) Percentage of methylcytosines in each sequence context in E16.5mPGC.











