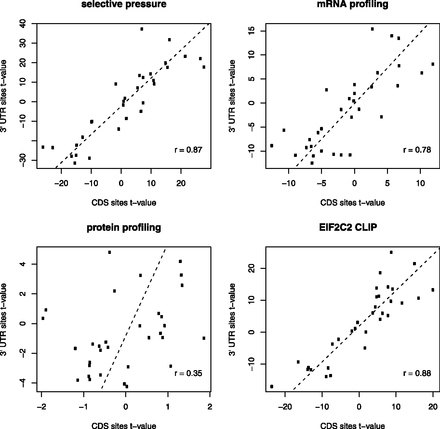
CDS and 3′ UTR sites share common sequence and structure properties. Sets of functional and nonfunctional binding sites were defined according to four different criteria—selective pressure, efficacy in mRNA degradation, efficacy in reducing protein levels, and binding to EIF2C2—each corresponding to a different panel. We then compared t-values obtained in comparing functional and nonfunctional sites from CDS (x-axis) and 3′ UTR (y-axis) regions. Each property is represented in each plot as a dot. Positive and negative values denote positive and negative predictors of functional miRNA binding sites, respectively.











