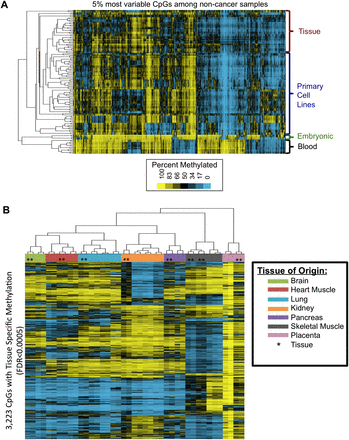
Noncancer samples exhibit methylation differences associated with cell culture, as well as tissue-specific methylation that is preserved between primary cell lines and tissues. (A) Unsupervised hierarchical clustering of the top 5% of CpGs with the most varying methylation across noncancer samples separates clades of samples characterized as tissues, primary cell lines, embryonic cell types, and blood leukocytes. The tissues and primary cell lines are divided into separate clades by a cell culture-associated methylation signature. (B) Seven tissue types were represented by both primary cell lines and tissues in this data set (tissue types listed in legend). ANOVA identified 117,795 CpGs significantly associated with tissue of origin (FDR < 0.05). For this visualization, we performed unsupervised hierarchical clustering on the 3223 significant CpGs with the largest standard deviation of PM values across the samples (SD ≥ 26). Both primary cell lines and primary tissues share a common tissue-specific methylation pattern, and the heat map displays the methylation patterns associated with each tissue of origin. Many CpGs are partially methylated in the tissues (black = 50%) at loci where the cell lines are completely methylated (yellow = 100%), indicating that heterogeneity among the cell types comprising the tissues results in a dampened signal compared to the pure cell population isolated in a cultured cell line (tissues marked by *, cell lines unmarked).











