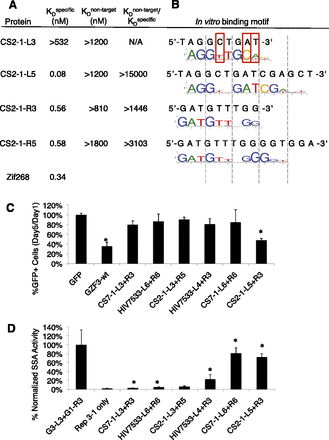
An analysis of affinity, specificity, and cytotoxicity. (A) EMSA was used to determine the affinity of CS2-1 three- and five-finger arrays for their specific (cognate) target as well as a nontarget (e.g., the L3 array on the R3 binding site). A large ratio of nontarget:specific binding is an indicator of good specificity. (B) In vitro binding specificity was also determined using the Bind-n-Seq target site selection assay (Zykovich et al. 2009). The binding preference of CS2-1-L3 appears to differ from the intended target site at three positions (red boxes). (C) Cytotoxicity was assessed as a decrease in the percentage of ZFN-expressing cells on day 5 compared to day 1. To follow only those cells expressing nuclease, a GFP expression vector was cotransfected with the indicated ZFN expression vectors. (GFP) Cells cotransfected with GFP and empty ZFN vector as a positive control. (GZF3-wt) A nuclease known to be cytotoxic (Szczepek et al. 2007) as a negative control. (Error bars) The standard deviation of normalized duplicates from at least two experiments. (*) P < 0.00001 compared to the GFP-only positive control based on a one-tailed homoscedastic t-test. (D) Cleavage activity is a summary of the SSA data from other figures and is shown here for reference.











