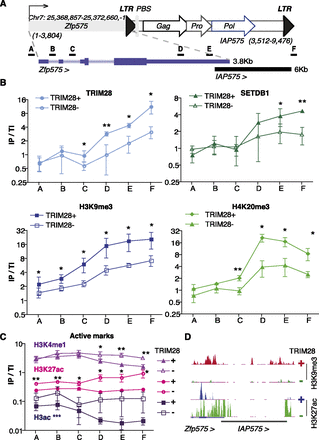
Zfp575 is regulated by a gain of active chromatin marks at its adjacent IAP575. (A) Map of Zfp575 and its adjacent IAP575 (for details, see Fig. 3A) with an enlargement shown underneath to show where primer pairs for ChIP are located. (B) ChIP results of repressive marks. (IP/TI) Immunoprecipitate values were normalized to their respective total inputs and to Gapdh. Bars represent the mean and SD of three to four ChIPs per antibody, and experiments were also reproduced in another ES cell line (Rex1) (data not shown). In each experiment, controls of no antibody were included giving no enrichments. Differences between WT and TRIM28-depleted samples were assessed for each primer set using paired t-tests with all significant differences given; (*) P ≤ 0.05, (**) P ≤ 0.01. (C) ChIPs this time on active marks were performed as described in B with data representing three to four ChIPs per antibody. Additionally, here the Pou5f1 enhancer was used as a positive control (data not shown) showing high enrichment for both H3K4me1 and H3K27ac but not for TRIM28 or H3K9me3. For H3K4me1 and H3K27ac, all significant differences are shown for each primer set, while for H3ac, WT samples were significantly different from TRIM28-depleted ones, not for individual points but over all primer sets; (***) P ≤ 0.001. (D) ChIP-seq maps of H3K9me3 and H3K27ac in TRIM28 WT and depleted ES cells (set to the same vertical scale) at the Zfp575-IAP575 locus. Note that reads within ERVs, especially conserved ones (in black), are usually missing due to the inability to map reads within highly repeated sequences. However, reads are present at the borders of these elements.











