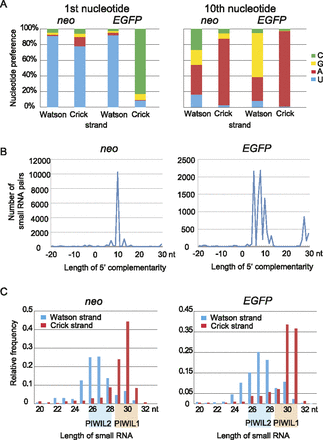
Small RNAs from EGFP-neo have features of piRNA. (A) Nucleotide preference at the first and 10th positions. Note that Watson strand small RNAs are enriched for U at the first position (a feature of the primary piRNA), and Crick strand small RNAs for A at the 10th position (a feature of the secondary piRNA). (B) Frequent 10-nt complementarity between the 5′ regions of Watson and Crick strand piRNAs. Small RNA pairs with 5′ complementarity for indicated lengths are counted. If eight reads of Watson strand small RNA and five reads of Crick strand small RNA had the same complementary sequences, it was counted as 8 × 5 = 40. The prevailing 10-nt complementarity suggests an active ping-pong cycle. (C) Size distributions of small RNAs from different strands. The Watson strand piRNAs are 25–28 nt in length and presumably bound by PIWIL2, and the Crick strand piRNAs are 29–32 nt in length and presumably bound by PIWIL1.











