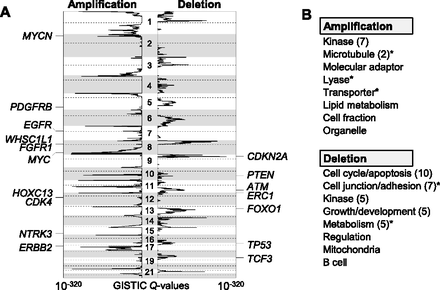
Recurrent chromosomal alterations across diverse tumor types. (A) The significance of recurrent amplification (left) and deletion (right) as measured by the GISTIC algorithm (GISTIC Q value; log-scaled) is plotted across the genome. Seventeen known targets (cancer consensus genes) are shown at the corresponding peaks. (B) The GO categories significantly enriched in pan-lineage amplification and deletion MCRs are shown. Similar functional categories (e.g., “kinase” or “kinase activity”) are grouped, and the representative function is shown with the number of related GO functions in parentheses. An asterisk indicates that the corresponding functional categories remained significant after removal of MCRs with known cancer genes.











