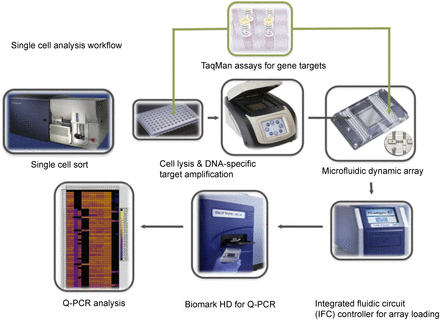
Schematic workflow of the multiplex targeted Q-PCR approach for the simultaneous detection of gene fusions, DNA copy number alterations, and mutations in single cells. Initially, CFSE-labeled single cells are sorted into each well of a 96-well plate and then lysed. Multiplex DNA specific target amplification is then completed to amplify a DNA region of interest (fusion gene, copy number alteration, or SNV) from the two copies found in a single cell to an amount that can be detected by Q-PCR using the BioMark HD. Amplified samples and assays are then loaded into a 96.96 dynamic array that utilizes a valve-controlled capillary network to bring these two mixes together at nanoliter volumes (completed in the IFC controller) for the Q-PCR reaction to take place. This final Q-PCR step determines the gene fusion, mutation, or copy number alteration status for each single cell.











