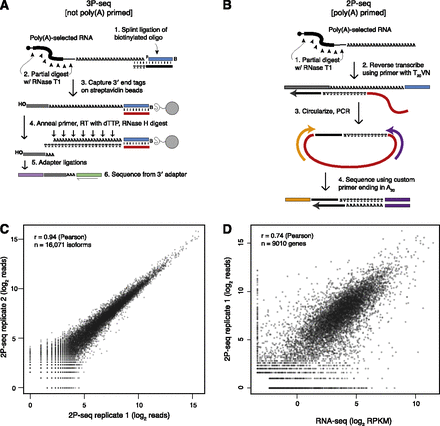
Experimental methods used to assay alternative cleavage and polyadenylation. (A) 3P-seq (Jan et al. 2011). This method captures 3′ end mRNA fragments without using oligo(dT) priming and, therefore, is highly specific for true poly(A) sites. A bridge oligo (black) is used to ligate a biotinylated oligo (blue); reverse transcription primer (red); captured mRNA 3′ end fragment (gray); Illumina sequencing adapters (purple and green). (B) Poly(A)-primed sequencing, or 2P-seq. This simplified method is useful for quantifying usage of known poly(A) sites. (C) Reproducibility of 2P-seq measurements of isoform abundance across two biological replicates. Only isoforms with at least a single read in either replicate were considered, and a single pseudocount was added prior to taking the log of each value. (D) Correspondence between mRNA quantified using 2P-seq and RNA-seq data sets from 3T3 cells. A minimum of two tags or 0.1 RPKM was required for 2P-seq and RNA-seq, respectively, and pseudocounts of 1 and 0.1, respectively, were added prior to taking the log of each value.











