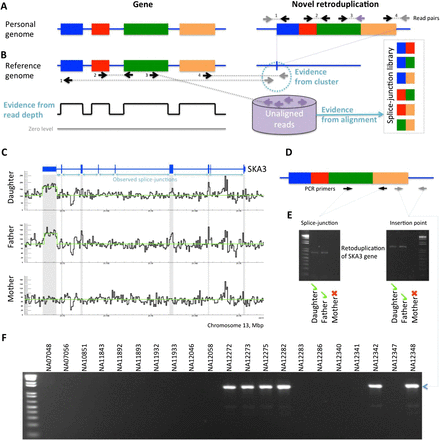
Approach for novel retroduplication discovery. (A) If an analyzed genome has an unknown (i.e., absent from reference genome) retroduplication, then sequencing reads originating from the retroduplication can be used for it to be discovered. (B) Reads aligned to the reference genome provide three lines of evidence for the novel retroduplication: reads clustering around the insertion point, increased read depth in exons, and mapping of unaligned reads to a splice-junction library. (C) The existence of a novel retroduplication for the SKA3 gene in the CEU trio is supported by the three lines of evidence. The retroduplication is polymorphic as it is not present in the mother's genome. (D) PCR validation strategy. Two sets of primers test for the presence of a splice junction and for the insertion point, respectively. (E) Existence of novel retroduplication for SKA3 is validated in the daughter's and father's genomes but not in the mother's. (F) The novel retroduplication for SKA3 is polymorphic in the CEU population, as PCR across the insertion point yields a product in only some of the individuals tested.











