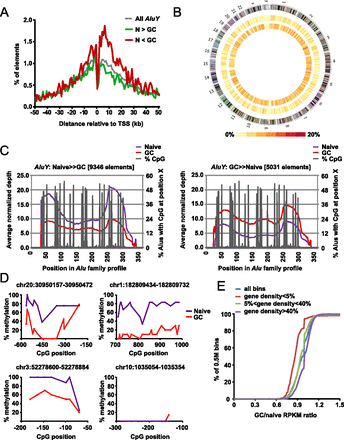
Distribution of differentially methylated Alus across the genome. (A) Distance of differentially methylated AluYs to the nearest TSS. The distribution of all AluY (dashed purple line) and both types of differentially methylated Alus, loss-of-methylation (N > GC) and gain-of-methylation (N < GC), are shown. (B) Circos plot (Krzywinski et al. 2009) illustrating the density of differentially methylated AluY elements (middle ring, GC > naive; inner ring, naive > GC) on each chromosome. Color density (from yellow to red) represents the percentage of AluY elements (from 0% to 20%) in each 5-Mb bin that is differentially methylated (in the given direction). (C) Average normalized depth of mapped reads from MIRA-seq across a consensus profile for AluY; the position in base pair is shown on the x-axis. Percentage of AluYs with CpG at each position is also shown. (Left) The average depth of AluYs that have lost methylation in GC B cells; (right) the average depth of AluYs that have gained methylation in GC B cells compared to naive B cells. (D) Analysis of methylation status of genomic regions neighboring differentially methylated Alus by genomic bisulfite sequencing. Regions proximal to four differentially methylated Alus in Figure 5C were analyzed. After bisulfite treatment and PCR amplification, at least eight individual clones of the PCR product were analyzed. The percentage of methylated cytosine (y-axis) was calculated at individual CpG positions relative to the 5′ end of each Alu element (x-axis). The genomic locations (hg19) of the Alu elements analyzed are listed above each graph. Methylation status of CpGs within individual PCR products are shown for five Alu elements in Supplemental Figure S4. (E) Overall DNA methylation dynamics at chromosomal regions with varying degrees of gene density between naive and GC B cells. We analyzed 5759 genomic bins of 0.5M bp for GC/naive adjusted RPKM ratios (there are a total of 6146 bins in the genome, but only those bins with GC content between 30% and 60% were analyzed). Gene density for a bin is defined as the fraction of positions that are within at least one RefSeq gene body. The data were partitioned into regions with low gene density (gene density < 5%, N = 1137, red line), intermediate gene density (5% < gene density < 40%, N = 1533, green line), and high gene density (gene density > 40%, N = 3100, purple line).











