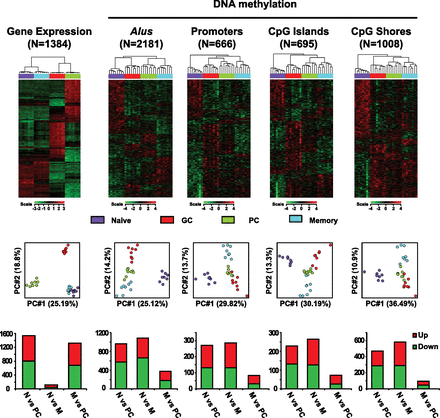
B-lymphocyte activation establishes DNA methylation reprogramming that poises memory B cells for recall responses. (Top) Heat maps displaying hierarchical clustering of genes differentially expressed (ANOVA P < 0.01 and fold change > 3) and genomic features with differential methylation scores (ANOVA P < 0.01). The total number of genes or genomic features is indicated. (Middle) PCA analysis of the corresponding differentially expressed genes or differentially methylated genomic features. Each circle represents an individual MIRA-chip sample; cell types are represented by different colors. x- and y-axes represent first and second principal components, respectively. The degree of variance in each principal component is indicated in parentheses. (Bottom) Analysis of genomic features by average score in naive B cells (N) vs. PC; naive B cells (N) vs. memory B cells (M); and memory B cells (M) vs. PC. The bar graph displays the number of genomic features with increased methylation (up) in red and decreased methylation (down) in green.











