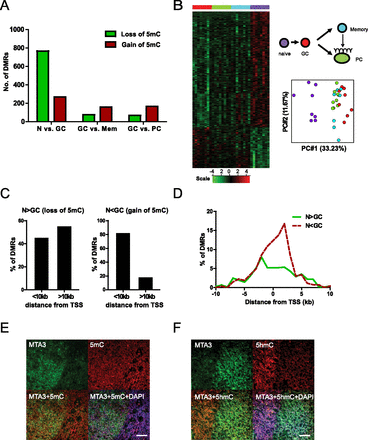
Widespread alteration of DNA methylation in B-cell subsets. (A) Assessment of global DNA methylation changes during immune activation (naive [N] vs. GC) and differentiation of GC B cells into memory or PCs (GC vs. memory [Mem] and GC vs. PC) by MIRA-chip analysis. The bar graph displays the number of DMRs with increased methylation (gain of 5mC) in red or decreased methylation (loss of 5mC) in green in each stage transition. (B) The methylation score is computed for each DMR between naive and GC B cells in all MIRA-chip samples and displayed in a heat map (left panel). Each row represents a DMR between naive and GC. Each column represents an individual MIRA-chip sample. All eight samples from each cell type clustered together in the heat map. Cell types are represented by different colors, as indicated in diagram beside the heat map. PCA analysis of methylation score of DMRs for all cell types is shown in the right panel. Each circle represents an individual biological sample. x- and y-axes represent first and second principal components, respectively. The degree of variance in each principal component is indicated in parentheses. (C) Frequency and genomic distribution of DMRs between naive and GC B cells. The bar graphs display the number of DMRs that are <10 kb or >10 kb from the nearest annotated TSS. The left panel represents DMRs with loss of methylation between naive and GC B cells (N > GC), while the right panel represents DMRs with gain of methylation between naive and GC B cells (N < GC). (D) Distribution of DMRs relative to the nearest TSS. The frequency of both types of DMRs, loss-of-methylation DMRs (N > GC), and gain-of-methylation DMRs (N < GC), in 1-kb increments from the TSS is shown in the graph. (E) Distribution of methylcytosine (5mC) in tonsils. A tonsil section was double stained for MTA3 to demarcate GC B lymphocytes (green) and 5mC (red). Nuclei were labeled with DAPI (blue). There was no detectable difference in the global level of 5mC in MTA3+ GC B cells and in non-GC B cells. Scale bar, 50 μm. (F) Distribution of hydroxy-methylcytosine (5hmC) in tonsils. A tonsil section was double stained for MTA3 to demarcate GC B lymphocytes (green) and 5hmC (red). Nuclei were labeled with DAPI (blue). The global level of 5hmC decreased in in MTA3+ GC B cells compared to non-GC B cells. Scale bar, 50 μm.











