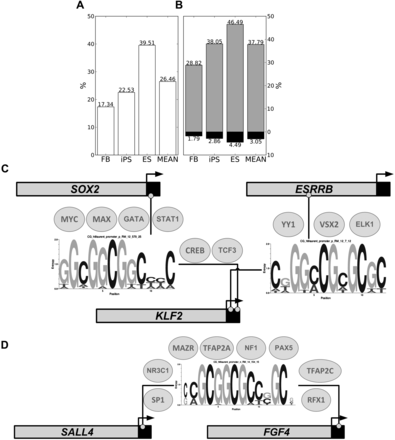
Crosstalk between CpGMMs and TFs. Percentages of CpGMM targets of each cell type (A) and methylation-resistant and methylation-prone CpGMM targets of each cell type (B) that co-occupy TFBSs. The methylation-resistant percentages are in gray, the methylation-prone in black, and the merged ones in white. The rightmost bars correspond to the mean across all populations. (C,D) Pluripotent networks arising from the crosstalk between CpGMMs and TFs. The CpGMMs are those whose loci targets have significant enrichment of pluripotent genes. The small circles indicate the positions of the methylation-resistant CpGMM target loci. The ellipses over the CpGMMs enclose the names of the TFs expressed >4.5 FPKM and whose TFBMs resemble (Pearson correlation ≥0.85) the CpGMM. The horizontal bars represent the promoters of the CpGMM targets. The light gray region of the bar represents the gene promoter itself; the black part represents a small portion of the coding region; and the beginning of each horizontal arrow marks the TSSs.











