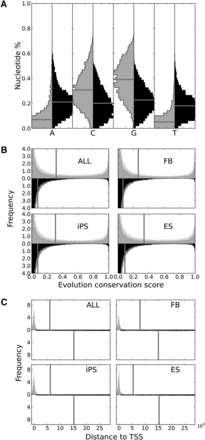
General discriminative features between all the methylation-resistant and methylation-prone CpGMMs. Histograms of (A) the 4 nt distributions across the discovered CpGMMs; (B) the conservation of all the CpGMM targets, where the sequence conservation scores were taken from primates phastCons 46-way (see Supplemental Material; Siepel et al. 2005); and (C) the distances of all the CpGMM targets to the TSSs. The methylation-resistant features are in gray; the methylation-prone, in black. Vertical lines mark the position of the median.











