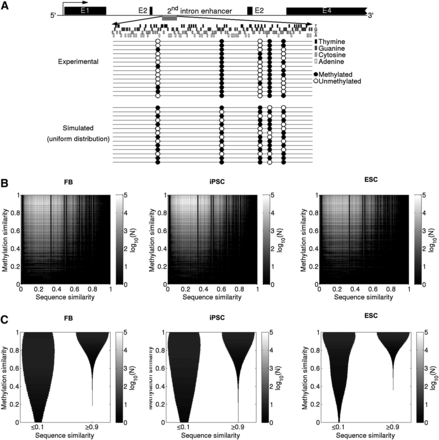
DNA methylation patterns are nonuniformly distributed and are influenced by their DNA context. (A) Analysis of the methylation state of nestin (NES) second intron enhancer region. The lollipop diagram in the top panel shows the observed 58.6% global methylation (Han et al. 2009). Such methylation is nonuniformly distributed along CpG “columns.” There are CpG “columns” with higher and lower probability to be methylated. In simulations with a uniform distribution of the methylation states, we can observe lollipop diagrams such as the one shown in the bottom panel, with the same methylation percentage as the one experimentally observed, but with nonpreferential “column” methylation distributions. (B) Heatmaps of the frequencies of the similarity of the methylation between the two DNA strands versus the similarity of the sequence of the two DNA strands for each CpG word. (C) Violin plots of the frequencies of the methylation similarity between the two DNA strands for low (≤0.1) and high (≥0.9) sequence similarity between the two DNA strands. The similarities are calculated genome wide, and their frequencies are represented in log10 scale by gray color bars.











