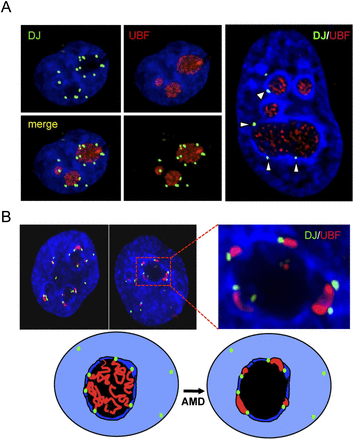
The DJ forms a perinucleolar anchor for rDNA repeats. (A) 3D immuno-FISH reveals that DJ sequences lie in perinucleolar heterochromatin in HT1080 cells. Nucleoli are visualized with UBF antibodies (red) and DJ with BAC CT476834 (green). The nucleus is DAPI-stained. The extended focus images (left) are stills from Supplemental Videos 1 and 2, while the image on the right shows a single focal plane. (B) Inhibition of rDNA transcription with AMD results in formation of nucleolar CAPs juxtaposed with DJ sequences in perinucleolar heterochromatin. Staining as in A. Two representative cells are shown, one with an enlargement. Cartoon models the transition between active and withdrawn rDNA upon AMD treatment. rDNA (red) retreats from the nucleolus (black) to the DJ (green) that is embedded in perinucleolar heterochromatin (dark blue).











