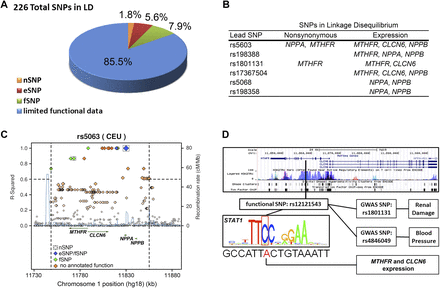
(A) Proportions of functional consequences of all SNPs in LD (r2 > 0.6) with the SNPs associated with BP and renal phenotypes at the AGTRAP-PLOD1 locus (see Supplemental Table 1). (B) Genes linked with lead SNPs of haplotypes associated with BP and renal phenotypes at the AGTRAP-PLOD1 locus. A lead SNP was considered linked to a gene if the lead SNP or SNPs in LD (r2 > 0.6) with a lead SNP caused nonsynonymous mutations or were significantly associated with expression of a gene. (C) An example of a BP-associated nSNP in NPPA (rs5063) that is in LD (r2 > 0.6) with an nSNP in MTHFR (rs2274976). Both are also in LD with fSNPs correlated with the expression of MTHFR, CLCN6, and NPPB. Note that a total of two nSNPs (boxed), two eSNPs (blue), and five fSNPs (green) are in LD with rs5063. (D) An example of an fSNP (rs12121543) that is in LD with two SNPs (rs1801131 and rs484609) associated with BP and renal phenotypes. An overview of the region between genes MTHFR and CLCN6 shows that rs12121543 falls in a transcriptional active region (vertical black bar overlapping histone modification, DNase hypersensitivity, and ChIP-seq peaks). rs12121543 is predicted to change a conserved nucleotide in a consensus STAT1-binding site and is associated with significant differences in MTHFR and CLCN6 expression compared with the major allele. (A,C,D) nSNP, nonsynonymous SNP; eSNP, expression SNP; and fSNP, functional SNP.











