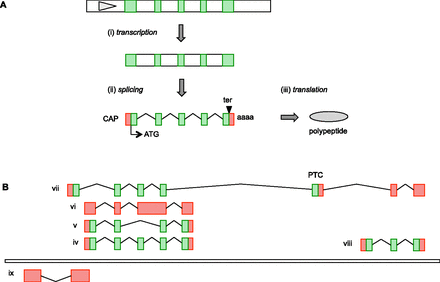
The evolving dogma of gene transcription. (A) The historical “central dogma” of molecular biology. By this model, (i) transcription generates the primary transcript (exons in green, introns in white), with the initial interaction between the RNA polymerase complex and the genome being mediated by a promoter region (gray triangle). (ii) The introns of the primary transcript are removed by the spliceosome, and a mature mRNA is generated by 5′ end capping (CAP) and polyadenylation (aaaa) (coding region [CDS] shown in green, untranslated 5′ and 3′ UTRs in red). (iii) The mRNA is translated into a polypeptide by the ribosome complex, with translation proceeding from the initiation codon (ATG) and ending at the termination codon (ter). (B) An updated model reflecting a modern view of transcriptional complexity. Here, the same gene (iv) undergoes alternative splicing (AS), for example an exon skipping event that does not change the frame of the CDS (v); this event thus has the potential to generate an alternative protein isoform. However, products of AS cannot be assumed to be functional; this gene has generated a retained intron transcript (vi), perhaps due to the failure of the spliceosome to remove this intron. Further complexity comes from a read-through transcription event (vii), whereby a transcript is generated that also includes exons from a neighboring protein-coding locus (viii). In this example, the read-through transcript has an alternative first exon compared with the upstream gene that contains a potential alternative ATG codon, although the presence of a subsequent premature termination codon (PTC) prior to two splice junctions indicates that this transcript is likely subjected to the nonsense mediated decay (NMD) degradation pathway. Finally, model ix is a transcript that is antisense to the upstream gene; both loci are potentially generated under the control of a bidirectional promoter.











