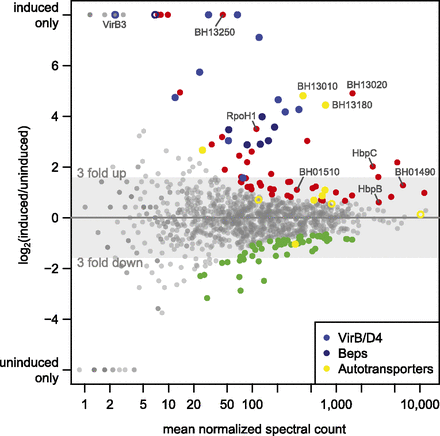
Differential protein expression analysis. The log2 fold change of the expression of all experimentally identified B. henselae proteins in the induced versus uninduced condition is shown against the mean normalized spectral count (MA plot). The 10% most significant differentially expressed proteins are highlighted, including 68 up-regulated proteins (red dots) and 57 down-regulated proteins (green dots). Selected regulated proteins are highlighted in different colors: members of the VirB/D4 T4SS (blue dots), Beps (dark blue dots), and several proteins containing autotransporter beta-domains (yellow dots). Proteins in these categories that rank below the 10% cutoff are shown as open circles.











