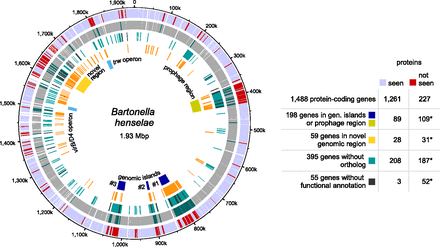
Integration of expression evidence with structural genome information and evolutionary conservation. Genes whose proteins were not identified cluster in specific regions of the B. henselae genome. Outer ring: Genes whose proteins were identified (light blue) or not identified (red). Second ring: Protein-coding genes classified by the RNA-seq analysis as expressed (gray) or not (dark green). Third ring: Genes without a detectable ortholog among species of lineage 4 of the genus Bartonella (Engel et al. 2011) (turquoise) and genes without any functional annotation by the eggNOG classification (black). Fourth ring: Repeat regions identified by RepSeek (orange) (Vallenet et al. 2006) and rRNA repeat regions (light orange). Fifth ring: Location of a prophage region (ochre), three genomic islands (blue), the virB/D4 and trw operons (sky blue), and a novel genomic region enriched in repeats as well as highly expressed genes whose encoded proteins were not identified (yellow). The results of hypergeometric tests for selected data sets are also shown (asterisks indicate statistically significant enrichment; see text). For the hypergeometric test, we used all possible protein-coding genes for the identified “seen” proteins (1250 distinct proteins encoded by 1261 gene models) and “not seen” proteins (217 distinct proteins encoded by 227 gene models). The circular plot was generated using DNAPlotter (Carver et al. 2009).











