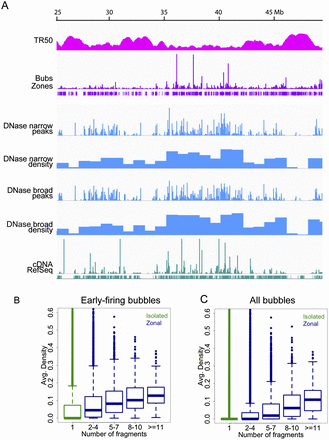
DNase I accessibility is strongly associated with origin activity in early-replicating chromatin. An IGV screen shot of a 25-Mb window of chromosome 22 displaying (from top to bottom) smoothed TR50 estimates over 50-kb windows in sliding steps of 1 kb, origin fragment RDs (bubbles), initiation zone calls (i.e., two or more adjacent bubble-containing fragments obeying the one-negative-fragment rule; see text), DNase I HSS read profiles at narrow-peak calls (Rep1), density of DNase I narrow-peak calls in 1-Mb windows (i.e., genomic coverage of the sites in base pairs divided by 1 Mb), DNase I HSS read profiles at broad-peak calls (Rep1), density of DNase I broad-peak calls in 1-Mb windows, transcript read profiles, and RefSeq gene annotations. (B,C) Boxplots of average DNase I HSS densities per bubble-containing fragment (i.e., genomic coverage of the sites across each EcoRI fragment divided by its length) as a function of origin fragment number, either in early-firing origins (B) or in all origins (C).











