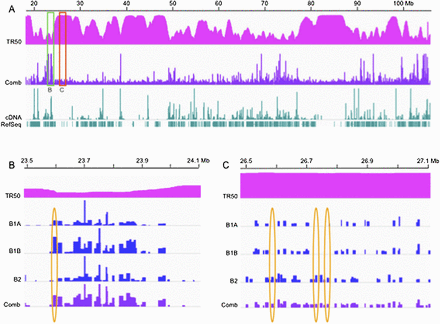
Origin distributions differ widely in both read depths (RDs) and reproducibility in different segments of the human genome. Integrative Genomics Viewer (IGV) screen shots of bubble-containing fragment RDs within a ∼90-Mb region of chr14. (A) TR50: time at which 50% of the DNA is replicated (scale: 0 to 6, representing the early-to-late-replication time windows defined in the text; Jeon et al. 2005). Comb: origin calls for the aggregate B1A, B1B, and B2 data sets (see text), where B1 and B2 correspond to biological replicate libraries, and B1A and B1B to sequencing replicates of the B1 library. Calls: bubble-containing EcoRI fragments with RDs greater than the respective cutoffs (0.0081 for B1A, 0.0139 for B1B, 0.0123 for B2, and 0.0144 for the combined set; Supplemental Table SI). (B) A 600-kb generally early-replicating region of chr14 corresponding to the region delimited by the green box in A. The scales of the IGV graphs were adjusted to aid comparison so that the highest RD in each data set corresponded to 100%, with the absolute range of values as follows: B1A, 0–0.273; B1B, 0–0.145; B2, 0–0.198; and Comb, 0–0.477. (C) A 600-kb late-replicating region of chr14 corresponding to the region outlined with the red rectangle in A. This IGV view has been expanded vertically by 2.5-fold to aid comparison of the four different data sets. The absolute scales remain the same except for this correction.











