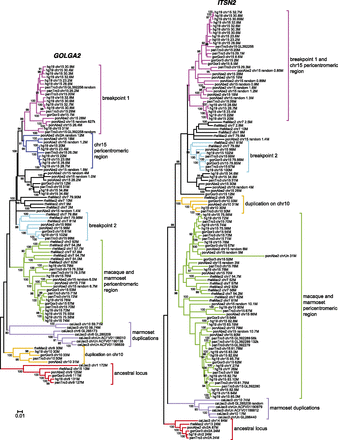
Primate phylogeny of GOLGA2 and ITSN2 duplicons. Neighbor-joining phylogenetic trees were constructed using a 2.5-kb region of the GOLGA2 module and a 3.3-kb region of the ITSN2 module. Sequences were retrieved from human (hg19), chimpanzee (panTro3), gorilla (gorGor3), orangutan (ponAbe2), macaque (rheMac2), and marmoset (calJac3) reference genomes. Trees are drawn to scale, with branch lengths measured in the number of substitutions per site and bootstrap values (1000 replicates) shown. Monophyletic clades of duplicons mapping to the ancestral loci (9q34.11 and 2p23.3 for GOLGA2 and ITSN2, respectively), chromosome 10, marmoset duplications, macaque and marmoset pericentromeric region, inversion breakpoints, and the pericentromeric region of ape chromosome 15 are highlighted in color code. The copies at the proximal inversion breakpoint and ape pericentromeric region homogenized, especially in the ITSN2 phylogenetic tree, do not group into two different branches.











