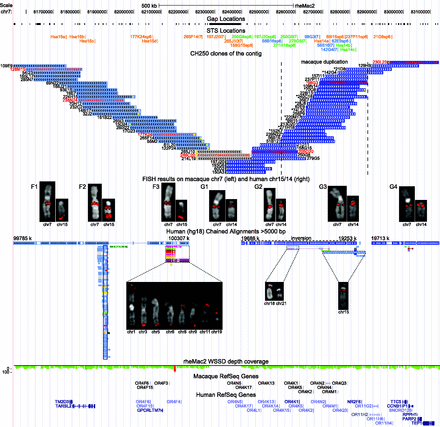
Contig of CH250 BAC clones on MMU7. The schematic shows features of the macaque genomic region spanned by the contig (chr7:81552000–83132000, rheMac2). In particular, gap and STS locations, BAC clones, FISH mapping data, chained alignments with hg18 reference, WSSD (whole-genome shotgun sequence detection) depth-of-coverage data (the green color corresponds to single copy; the red color to duplication; Marques-Bonet et al. 2009), macaque RefSeq genes, and human RefSeq genes in the orthologous human regions are shown. STS names are color-coded: STS used in radioactive hybridization (orange), STS used in PCR amplification (blue), and STS used in both assays (green). FISH results on MMU7 and human chromosomes 14/15 are shown for representative BAC clones marked by names and arrowheads in red. Seven different FISH patterns are identified: BAC clones of Groups F1–F3 map to the homologous region on chromosome 15 (light blue); BAC clones of Groups G2–G4 map to the homologous region on chromosome 14 (dark blue); and gray-colored BAC clones (Group G1) showed absent or weak signal on human chromosome 14 and correspond to the fission breakpoint. An asterisk in front of the BAC name indicates BAC clones belonging to the second copy of the macaque duplication. A yellow arrowhead marks BES not anchored to the human reference hg18. Additional FISH signals on other human chromosomes due to the presence of interchromosomal SDs (further indicated by the chained alignments) are shown for the BAC clones of Groups F3, G2, and G3. An empty black box marks the human GOLGA2–ITSN2 core duplicon. Both end segments of the contig (Groups F1 and G4) are in single copy and direct orientation in macaque and human.











