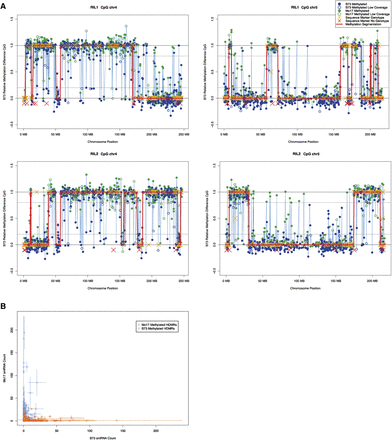
Inheritance and stability of DNA methylation patterns. (A) 9635 regions in the genome where B73 and Mo17 have different CG methylation levels were found using a moving window and a χ2 test based on C and T counts at CG sites. The regions were chosen so that one parent is <10% methylated and the other >70%. The regions are at least 200 bp long and have at least 20 CG sites. There is one point on the plots for each of these 9635 regions. The x-axis is the region number in chromosome order. The y-axis value is (%C in RIL − %C in B73)/(%C in Mo17 − %C in B73). Thus, data points approaching 1 are similar to Mo17, and those approaching 0 to B73 in methylation level. Mapped (orange) and unmapped (red) markers indicate recombination break points. (B) Scatterplots of small RNA levels that match HDMR in each inbred. Error bars represent variation in small RNA levels found in three biological replicates.











