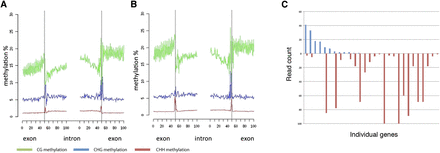
Splice site methylation. (A) Metagene analysis of splice junctions on the antisense strand, in CG (green), CHG (blue), and CHH (red) contexts. (B) Metagene analysis of splice junctions on the sense strand, in CG (green), CHG (blue), and CHH (red) contexts. (C). Alternatively spliced genes (n = 28) analyzed for CHG methylation levels at alternative acceptor sites (Supplemental Table S7B). The number of RNA-seq reads is plotted at alternate acceptor sites with high (blue, above the line) and low (red, below the line) methylation for each individual gene.











