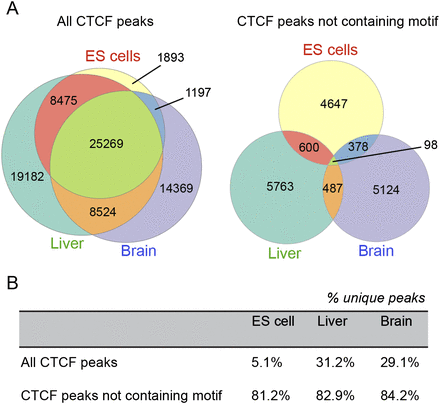
(A) Proportional Venn diagrams comparing coincidence of CTCF binding sites between ES cells, liver, and brain demonstrate significant overlap of CTCF binding in these tissues, Coincident binding was also considered after the removal of binding regions containing the consensus CTCF motif; overlap of CTCF binding in the absence of the consensus motif was much lower than when all binding sites were considered. (B) The percentages of shared peaks for each tissue type for all peaks and for nonmotif peaks.











